Dr. rer. nat. Stefan Kempa

Integrative Metabolomics and Proteomics
Max-Delbrück Center for Molecular Medicine (MDC)
Robert-Rössle-Str. 10
13092 Berlin
Tel: +49 30 9406-3033
E-Mail: stefan.kempa(at)mdc-berlin.de
https://www.mdc-berlin.de/proteomics-metabolomics
University Education
- 1995-2001 Studies of Biochemistry, Universität Potsdam
- 2001-2004 Ph.D., Max Planck Institute for Molecular Plant Pharmacology
Professional Experience (selection)
- 2004-2007 Postdoctoral research fellow and independent researcher at the Institute for Molecular Biotechnology, IMBA/GMI Vienna, Austria
- 2007-2009 Senior postdoctoral research fellow at the Universität Potsdam, GoForSys Project
- Since 2009 Group leader, integrative metabolomics and proteomics, MDC
Scientific Scope
Within the past decades biochemical data of single processes, metabolic and signaling pathways were collected and advances in technology led to improvements of sensitivity and resolution of bioanalytical techniques. These achievements build the bases for the so called ‘genome wide biochemistry’. In this context the aim of our group is to apply metabolomics and proteomics techniques for absolute quantification and to analyze turnover rates of proteins and metabolites using stable isotopes. Furthermore we analyze posttranslational modifications of proteins e.g. by quantitative phospho-proteomics.
Our core interest is the characterization of post-transcriptional and post-translational regulatory mechanisms which play an important role in metabolic regulation. Therefore we have developed new techniques to measure the metabolic fluxes within the central metabolism using stable isotope labeled substrates e.g. glucose, glutamine, pyruvate or palmitate and combine this technology with state of the art proteomics methods. Using these techniques we could analyze the translational regulation of an RNA binding protein (Jungkamp et al. 2011) in vivo and could resolve the mode of action of a synthetic lethal interaction between MYC and the protein kinase ARK5 (Liu et al. 2012).
In frame of future projects we will apply phospho-proteomics, in combination with pulsed stable isotope resolved metabolomics (pSIRM) to identify new targets of metabolic regulation in cancer cells.
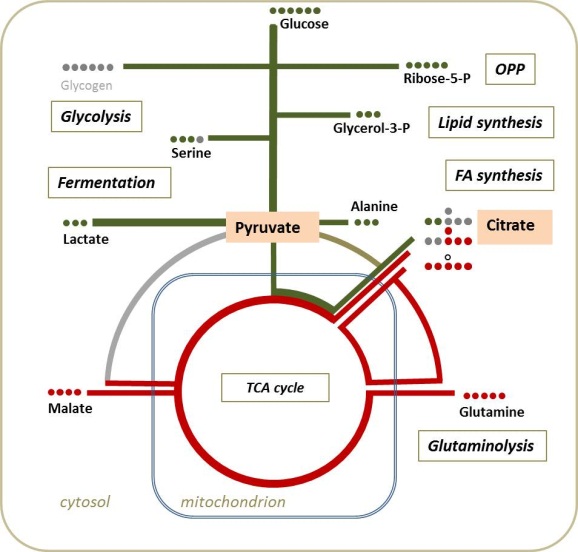
Scheme of the central metabolism of cancer cells
Distribution of isotopomers resulting from pulsed stable isotope resolved metabolomics after labeling with 13C-glucose or 13C-glutamine.
Selected References
- Dal Santo, S., Stampfl, H., Krasensky, J., Kempa, S., Gibon, Y., Petutschnig, E., Rozhon, W., Heuck, A., Clausen, T., Jonak, C.(2012) "Stress-Induced GSK3 Regulates the Redox Stress Response by Phosphorylating Glucose-6-Phosphate Dehydrogenase in Arabidopsis." Plant Cell. 2012 Aug 10.
- Mastrobuoni, G., Irgang, S., Pietzke, M., Wenzel, M., Assmus, H.E., Schulze, W.X., Kempa, S.(2012) "Proteome dynamics and early salt stress response of the photosynthetic organism Chlamydomonas reinhardtii." BMC Genomics. 2012 May 31;13(1):215.
- Onal, P., Grün, D., Adamidi, C., Rybak, A., Solana, J., Mastrobuoni, G., Wang, Y., Rahn, H.P., Chen, W., Kempa, S., Ziebold, U., Rajewsky, N.(2012) "Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells." EMBO J. 2012 Apr 27;31(12):2755-69.
- Liu, L., Ulbrich, J., Müller, J., Wüstefeld, T., Aeberhard, L., Kress, T.R., Muthalagu, N., Rycak, L., Rudalska, R., Moll, R., Kempa, S.*, Zender, L., Eilers, M., Murphy, D.J. (2012) "Deregulated MYC expression induces dependence upon AMPK-related kinase 5." Nature. 2012 Mar 28;483(7391):608-12. *Corresponding for metabolomics and proteomics
- Jungkamp, A.C., Stoeckius, M., Mecenas, D., Grün, D., Mastrobuoni, G., Kempa, S., Rajewsky, N.(2011) "In vivo and transcriptome-wide identification of RNA binding protein target sites." Mol Cell. 2011 Dec 9;44(5):828-40.
- Adamidi, C., Wang, Y., Gruen, D., Mastrobuoni, G., You, X., Tolle, D., Dodt, M., Mackowiak, S.D., Gogol-Doering, A., Oenal, P., Rybak, A., Ross, E., Sanchez Alvarado, A., Kempa, S.*, Dieterich, C., Rajewsky, N., Chen, W. (2011) "De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics." Genome Res. 2011 Jul;21(7):1193-200. *Co-corresponding
- Kempa, S., Hummel, J., Schwemmer, T., Pietzke, M., Strehmel, N., Wienkoop, S., Kopka, J., Weckwerth, W.(2009) "An automated GCxGC-TOF-MS protocol for batch-wise extraction and alignment of mass isotopomer matrixes from differential 13C-labelling experiments: a case study for photoautotrophic-mixotrophic grown Chlamydomonas reinhardtii cells." J Basic Microbiol. 2009 Feb;49(1):82-91. PubMed PMID: 19206143.
- May, P., Wienkoop, S., Kempa, S.*, Usadel, B., Christian, N., Rupprecht, J., Weiss, J., Recuenco-Munoz, L., Ebenhöh, O., Weckwerth, W., Walther, D.(2008) "Metabolomics- and proteomics-assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii." Genetics. 2008 May;179(1):157-66. * Co-first author
- Kempa, S., Krasensky, J., Dal Santo, S., Kopka, J., Jonak, C.(2008) "A central role of abscisic acid in stress-regulated carbohydrate metabolism." PLoS One. 2008;3(12):e3935. Epub 2008 Dec 12. PubMed PMID: 19081841; PubMed Central PMCID: PMC2593778.
- Kempa, S., Rozhon, W., Samaj, J., Erban, A., Baluska, F., Becker, T., Haselmayer, J., Schleiff, E., Kopka, J., Hirt, H., Jonak, C. (2007) "A plastid-localized glycogen synthase kinase 3 (GSK3) modulates stress tolerance and carbohydrate metabolism." Plant J. 2007 Mar;49(6):1076-90.



